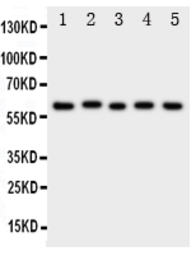
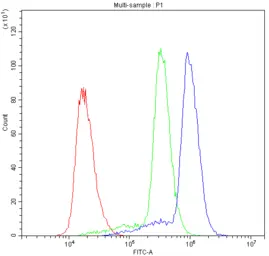
ASIC3 antibody
Cat. No. GTX01177
Cat. No. GTX01177
-
HostRabbit
-
ClonalityPolyclonal
-
IsotypeIgG
-
ApplicationsWB FCM
-
ReactivityHuman, Mouse, Rat