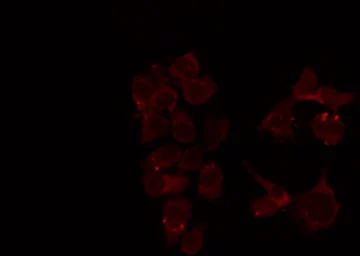
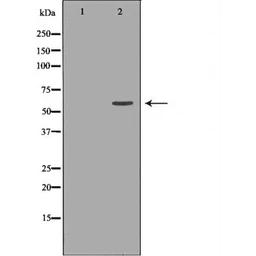
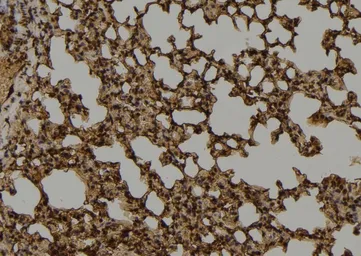
MUTYH antibody
Cat. No. GTX03359
Cat. No. GTX03359
-
HostRabbit
-
ClonalityPolyclonal
-
IsotypeIgG
-
ApplicationsWB ICC/IF IHC-P
-
ReactivityHuman, Mouse